|
Size: 9553
Comment:
|
Size: 9742
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 8: | Line 8: |
| * Genomics technologies are very sensitive: they can detect small amounts of variation. A good experimental design considers all possible variables (or factors) and ensures better quality data. You are encouraged to come to talk with us (and/or with the biostatistician Shaheena Bashir) about your planned experiment. Will your design enable you to answer your questions? Are there variables you did not think of? Do you have enough replicates? Is your control appropriate for downstream analysis? * These consulting meetings can also generate a follow-up plan, where additional meetings can be scheduled during and after an experiment is run to answer questions and check that the experimental design is compatible with downstream analysis: it is up to you to decide if you need the analysis or not. We are always available to discuss your data. |
* Genomics technologies are very sensitive: they can detect small amounts of variation. A good experimental design considers all possible variables (or factors) and ensures better quality data. You are encouraged to come to talk with us (and/or with the biostatistician Shaheena Bashir, sbashir@uhnres.utoronto.ca) about your planned experiment. '''''Will your design enable you to answer your questions'''''? Are there variables you did not think of? Do you have enough replicates? Is your control appropriate for '''''downstream analysis'''''? * These consulting meetings can also generate a follow-up plan, where additional meetings can be scheduled during and after an experiment is run to answer questions and check that the experimental design is compatible with '''''downstream analysis''''': it is up to you to decide if you need the analysis or not. We are always available to discuss your data. |
| Line 13: | Line 13: |
| * ~+'''Goal'''+~: To learn about your project and discuss pathway and network analysis options and their requirements. * Once correctly formatted input data are received and the quality is checked against analysis requirements, we will issue an '''initial pathway analysis plan''' (see below). * ~+'''time estimate'''+~: 30min to 1 hour * '''Initial Meeting and Data Input Requirement''': please have these data and information ready: * '''During the initial meeting, we will discuss:''' * the biological question(s) you want to answer * the experimental design * the platform you used to generate your data (e.g Affymetrix or Illumina, the chip model,...) * analysis already completed * the quality controls and the required input data format |
* ~+'''Goal'''+~: To learn about your project and discuss pathway and network analysis options and their requirements. * Once correctly formatted input data are received and the quality is checked against analysis requirements, we will issue an '''initial pathway analysis plan''' (see below). * ~+'''time estimate'''+~: 30 to 60 minutes * '''Initial Meeting and Data Input Requirement''': please have these data and information ready: * '''During the initial meeting, we will discuss:''' * the biological question(s) you want to answer * the experimental design * the platform you used to generate your data (e.g Affymetrix or Illumina, the chip model,...) * analysis already completed * the quality controls and the required input data format |
| Line 24: | Line 24: |
| * '''Your data should have been statistically analyzed''' (you should provide us with one file containing this information): * The data should have been normalized. * Some control quality plots should have been done: * Box-plot of intensity (before and after normalization) * Principal Component Analysis (PCA) * Hierarchical clustering of samples (performed on all the data) * Please provide a powerpoint presentation with a figure for each analysis |
* '''Your data should have been statistically analyzed''': * The data should have been normalized. * Some control quality plots should have been done: * Box-plot of intensity (before and after normalization) * Principal Component Analysis (PCA) * Hierarchical clustering of samples (performed on whole data) * Please provide a powerpoint presentation with a figure for each analysis |
| Line 32: | Line 32: |
| * An appropriate statistical test testing your hypothesis (your biological question) should have been performed, for example: moderated t-test, paired t-test, ANOVA, ... * If you need support for your statistical analyses, please contact Shaheena Bashir (Ph.D. in Statistics) at sbashir@uhnres.utoronto.ca. Located at MaRS TMDT 15th floor, she offers free consultation for statistical analyses for Cancer Stem Cell program (https://sites.google.com/site/biostatisticscancerstemcell/). She will analyze your data and output the results in the correct format for subsequent pathway and network analyses. You are encouraged to contact Shaheena as soon as you plan your experiment: these genomics technologies are very sensitive to noise and a well designed experiment is very important for best results. Statistical consultation at the design stage is crucial for improved data quality. |
* An appropriate statistical test testing your hypothesis (your biological question) should have been performed, for example: moderated t-test, paired t-test, ANOVA, ... * If you need support for your statistical analyses, please contact Shaheena Bashir (Ph.D. in Statistics) at sbashir@uhnres.utoronto.ca. Located at MaRS TMDT 15th floor, she offers free consultation for statistical analyses for Cancer Stem Cell program (https://sites.google.com/site/biostatisticscancerstemcell/). She will analyze your data and output the results in the correct format for subsequent pathway and network analyses. You are encouraged to contact Shaheena as soon as you plan your experiment: these genomics technologies are very sensitive to noise and a well designed experiment is very important for best results. Statistical consultation at the design stage is crucial for improved data quality. |
| Line 36: | Line 36: |
| * '''You need to provide us with 1 file (.txt) for enrichment analysis''' : * Name your file as follows: yourname_date.txt (example: veronique_March21.txt) * Please rename your file with a new date if you resubmit your file * Please follow the format description: * the first column corresponds to Entrez ID. * An Entrez ID is a numerical value that uniquely identifies genes. * For example the Entrez ID for Myc (myelocytomatosis oncogene [ Mus musculus ]) is 17869: http://www.ncbi.nlm.nih.gov/gene/17869. * the second column corresponds to a unique array identifier (ProbesetID for Affymetrix and sampleID for Illumina). * the third column corresponds to gene name (official gene symbol). * the fourth column corresponds to the gene description (full gene name). * the fifth and sixth columns contain the statistical values : * the statistical values are the ones that enable you to tell if a gene is significantly differentially expressed or not, it could be for example the t value and the p-value if you applied a t-test. * the whole table is ranked on the basis of adjusted p-value. * the additional columns contain the transformed (log2 for example) and normalized (RMA or quantile normalization for example) values for each sample (= each chip if gene expression data). |
* '''You need to provide us with 1 file (.txt) for enrichment analysis''' : * Name your file as follows: yourname_date_PIname.txt (example: veronique_March21_BADER.txt) * Please rename your file with a new date if you resubmit your file * Please follow the format description: * the first column corresponds to Entrez Gene ID. * An Entrez Gene ID is a numerical value that uniquely identifies genes. * For example the Entrez Gene ID for Myc (myelocytomatosis oncogene [ Mus musculus ]) is 17869: http://www.ncbi.nlm.nih.gov/gene/17869. * the second column corresponds to a unique array identifier (ProbesetID for Affymetrix and sampleID for Illumina). * the third column corresponds to gene name (official gene symbol). * the fourth column corresponds to the gene description (full gene name). * the fifth and sixth columns contain the statistical values : * the statistical values are the ones that enable you to tell if a gene is significantly differentially expressed or not, it could be for example the t value and the p-value if you applied a t-test. * the whole table is ranked on the basis of adjusted p-value. * the additional columns contain the transformed (log2 for example) and normalized (RMA or quantile normalization for example) values for each sample (= each chip if gene expression data). |
| Line 51: | Line 51: |
| * '''Example''': ||Entrez ID||Probeset ID||Gene Name||Gene Description||t value||p-value||sample1||sample2||sample3|| |
* '''Example''': ||Entrez ID||Probeset ID||Gene Name||Gene Description||t value||p value||sample1||sample2||sample3|| |
| Line 59: | Line 59: |
| * '''Note''': * Each row of the table should correspond to a different gene. If several rows correspond to the same gene (same Entrez ID), there are 2 possibilities to remove the redundancy: * for a same gene, only the row corresponding to the most extreme t-value is conserved * for a same gene, the average of the different normalized values is calculated before the t-test is applied * the choice must be made before the statistical data analyses are performed. We can discuss it during the initial meeting. * Include all your data (even data with non significant p-values) |
* '''Note''': * Each row of the table should correspond to a different gene. If several rows correspond to the same gene (same Entrez ID), there are 2 possibilities to remove the redundancy: * for a same gene, only the row corresponding to the most extreme t-value is conserved * for a same gene, the average of the different normalized values is calculated before the t-test is applied * the choice must be made before the statistical data analyses are performed. We can discuss it during the initial meeting. * Include all your data (even data with non significant p-values) * a web tool to facilitate the conversion from different identifiers (e.g. gene symbol, probsetID) to Entrez Gene IDs: THE SYNERGIZER (http://llama.mshri.on.ca/synergizer/translate/) |
Service SOP
Consulting meeting
Goal: to help plan an experiment before it is run. We can recommend case studies that user can learn from.
Genomics technologies are very sensitive: they can detect small amounts of variation. A good experimental design considers all possible variables (or factors) and ensures better quality data. You are encouraged to come to talk with us (and/or with the biostatistician Shaheena Bashir, sbashir@uhnres.utoronto.ca) about your planned experiment. Will your design enable you to answer your questions? Are there variables you did not think of? Do you have enough replicates? Is your control appropriate for downstream analysis?
These consulting meetings can also generate a follow-up plan, where additional meetings can be scheduled during and after an experiment is run to answer questions and check that the experimental design is compatible with downstream analysis: it is up to you to decide if you need the analysis or not. We are always available to discuss your data.
Analysis planning meeting
Goal: To learn about your project and discuss pathway and network analysis options and their requirements.
Once correctly formatted input data are received and the quality is checked against analysis requirements, we will issue an initial pathway analysis plan (see below).
time estimate: 30 to 60 minutes
Initial Meeting and Data Input Requirement: please have these data and information ready:
During the initial meeting, we will discuss:
- the biological question(s) you want to answer
- the experimental design
- the platform you used to generate your data (e.g Affymetrix or Illumina, the chip model,...)
- analysis already completed
- the quality controls and the required input data format
Your data should have been statistically analyzed:
- The data should have been normalized.
- Some control quality plots should have been done:
- Box-plot of intensity (before and after normalization)
- Principal Component Analysis (PCA)
- Hierarchical clustering of samples (performed on whole data)
- Please provide a powerpoint presentation with a figure for each analysis
- An appropriate statistical test testing your hypothesis (your biological question) should have been performed, for example: moderated t-test, paired t-test, ANOVA, ...
If you need support for your statistical analyses, please contact Shaheena Bashir (Ph.D. in Statistics) at sbashir@uhnres.utoronto.ca. Located at MaRS TMDT 15th floor, she offers free consultation for statistical analyses for Cancer Stem Cell program (https://sites.google.com/site/biostatisticscancerstemcell/). She will analyze your data and output the results in the correct format for subsequent pathway and network analyses. You are encouraged to contact Shaheena as soon as you plan your experiment: these genomics technologies are very sensitive to noise and a well designed experiment is very important for best results. Statistical consultation at the design stage is crucial for improved data quality.
You need to provide us with 1 file (.txt) for enrichment analysis :
- Name your file as follows: yourname_date_PIname.txt (example: veronique_March21_BADER.txt)
- Please rename your file with a new date if you resubmit your file
- Please follow the format description:
- the first column corresponds to Entrez Gene ID.
- An Entrez Gene ID is a numerical value that uniquely identifies genes.
For example the Entrez Gene ID for Myc (myelocytomatosis oncogene [ Mus musculus ]) is 17869: http://www.ncbi.nlm.nih.gov/gene/17869.
- the second column corresponds to a unique array identifier (ProbesetID for Affymetrix and sampleID for Illumina).
- the third column corresponds to gene name (official gene symbol).
- the fourth column corresponds to the gene description (full gene name).
- the fifth and sixth columns contain the statistical values :
- the statistical values are the ones that enable you to tell if a gene is significantly differentially expressed or not, it could be for example the t value and the p-value if you applied a t-test.
- the whole table is ranked on the basis of adjusted p-value.
- the additional columns contain the transformed (log2 for example) and normalized (RMA or quantile normalization for example) values for each sample (= each chip if gene expression data).
- the first column corresponds to Entrez Gene ID.
Example:
Entrez ID |
Probeset ID |
Gene Name |
Gene Description |
t value |
p value |
sample1 |
sample2 |
sample3 |
17218 |
10572906 |
Mcm5 |
minichromosome maintenance deficient 5, cell division cycle |
44.0079 |
0.001 |
9.13084 |
9.7166 |
8.76638 |
27279 |
10448307 |
Tnfrsf12a |
tumor necrosis factor receptor superfamily, member 12a |
-41.815 |
0.001 |
8.58977 |
9.29698 |
8.80844 |
13215 |
10582809 |
Tk1 |
thymidine kinase 1 |
39.9456 |
0.001 |
8.94519 |
9.56513 |
8.38612 |
12937 |
10384145 |
H2afv |
H2A histone family, member V |
-33.6475 |
0.001 |
10.574 |
10.7741 |
10.5401 |
207277 |
10526848 |
33.3352 |
0.001 |
8.25088 |
8.4121 |
8.2783 |
Note:
- Each row of the table should correspond to a different gene. If several rows correspond to the same gene (same Entrez ID), there are 2 possibilities to remove the redundancy:
- for a same gene, only the row corresponding to the most extreme t-value is conserved
- for a same gene, the average of the different normalized values is calculated before the t-test is applied
- the choice must be made before the statistical data analyses are performed. We can discuss it during the initial meeting.
- Include all your data (even data with non significant p-values)
a web tool to facilitate the conversion from different identifiers (e.g. gene symbol, probsetID) to Entrez Gene IDs: THE SYNERGIZER (http://llama.mshri.on.ca/synergizer/translate/)
- Each row of the table should correspond to a different gene. If several rows correspond to the same gene (same Entrez ID), there are 2 possibilities to remove the redundancy:
Analysis
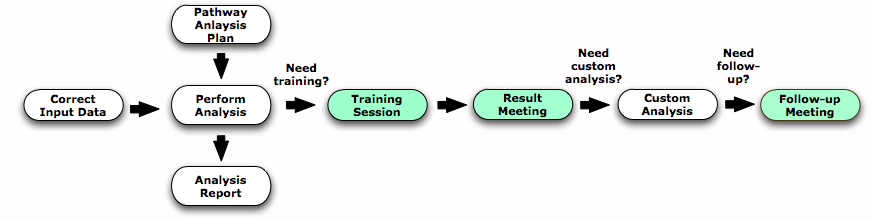
Pathway Analysis Plan
Goal: A pathway analysis plan is a document that states the different analyses that will be performed and a completion time estimate. We write the pathway analysis plan once correctly formatted input data are received. It needs to be signed off by researchers leading the project and the lead P.I.
- A meeting can be scheduled if requested to explain the Pathway Analysis Plan.
Run analysis, interpret the results and produce a report
Status : the analysis status is visible on the website page (see at the end of the page); We will communicate with you very regularly during the process to ensure effective interpretation of results.
Analysis Report: A report will include an overview of the results and a detailed focus analysis of interesting pathways will be written at the end of the analysis.
Result Meeting
- Goal: discuss the analysis and report.
- Examples of questions we can discuss: Do the results meet your expectations?
- Is there anything unexpected in the results? If you had the resources, which experiments would you conduct based on the results of this analysis?
- Examples of questions we can discuss: Do the results meet your expectations?
- Time estimate: 30min to 1hour
- Two options are available after this meeting:
- We need to perform additional bioinformatics analyses: customized analyses
- You are satisfied with the results and you explore the data and results using available software tools that we provide and then perform some validation experiments before an optional follow-up meeting
- Goal: discuss the analysis and report.
Training session
Goal: You can schedule a training session if you wish to do your own pathway and network analysis or explore results we have generated. We will explain how to install the required software and how to use it to explore your data.
time estimate: 30min to 1 hour
Customized analyses
- Meeting with Researcher to explain the results of the customized analyses
Follow-up
- Goal: you may have performed validation experiments or generated new research hypotheses based on your genomics study. You may need to go back and focus on a different aspect of your data. We can help you to re-analyse your data, provide additional bioinformatics tools or help plan a subsequent genomics experiment.
List of projects
- This section summarizes the current projects, and the analysis status for each project. You can see progress in the analysis of your project and see the different priorities assigned to each project.
project |
lab |
data received |
data checked; OK for analysis |
pathway analysis plan |
GSEA |
First Map |
Analysis report |
additional analysis |
status |
priority |
EZ01 |
Zacksenhaus |
Feb 22 |
Feb 23 |
Feb 24 |
Feb 25 |
Feb 26 |
March 30 |
report shared |
on-going |
1 |
JD02 |
Dick |
March 24 |
March 29 |
- |
- |
- |
- |
- |
- |
? |
JD03 |
Dick |
- |
- |
- |
- |
- |
- |
- |
- |
? |
JD04 |
Dick |
- |
- |
- |
- |
- |
- |
- |
- |
? |
CD02 |
Guidos |
- |
- |
- |
- |
- |
- |
- |
- |
? |
