|
Size: 1408
Comment:
|
Size: 5375
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| #acl CscGroup:read,write,revert | ## page was renamed from CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService #acl All:read <<BR>> = OICR Cancer Stem Cell program - Pathway and Network Analysis Service = <<BR>> |
| Line 3: | Line 7: |
| = Pathway and Network Analysis Service = | * '''The Pathway and Network Analysis Service''' is freely available to all Cancer Stem Cell program members. <<BR>> * '''High-throughput genomic experiments''' (e.g. gene expression, large-scale genetic screens) often lead to the identification of large gene lists. The interpretation of results and the formulation of consistent biological hypotheses from these gene lists can be challenging. Pathway and network analysis (e.g enrichment analysis) approaches can aid interpretation by relating the gene list to knowledge about the biological system, such as pathways. |
| Line 5: | Line 11: |
| == Cancer Stem Cell program == | * '''Our goal''' is to help researchers interpret results of genomics experiments. Analysis is conducted in close collaboration with researchers on each project to ensure correct input data and effective interpretation of results. Ideally researchers do as much of the analysis and interpretation as they can. |
| Line 7: | Line 13: |
| ---- ----- |
* '''Standard types of pathway analysis offered''' * '''Pathway and network analysis: find pathways enriched in a list of genes (e.g. differentially expressed genes)''' * Gene-set enrichment analysis helps characterize large gene lists by finding functionally coherent gene-sets, such as pathways, that are statistically over-represented in a given gene list. We have also developed a method to visualize the results of this analysis, called Enrichment Map. Enrichment Map organizes gene-sets in a network and it enables the user to quickly identify the major enriched functional themes. '''Input''': gene list from genomics experiment (statistically analyzed). '''Output''': enriched pathways visually displayed. * Related publications: [[http://www.ncbi.nlm.nih.gov/pubmed/16199517|Gene-set enrichment analysis (GSEA)]] [[http://www.ncbi.nlm.nih.gov/pubmed/21085593|Enrichment Map]] * '''Predict the function of an unknown gene''' GeneMANIA finds other genes that are related to a set of input genes, using a very large set of functional association data. '''Input''': a gene or set of genes. '''Output''': connections between input genes and suggestions for additional related genes. * Related publications: [[http://www.ncbi.nlm.nih.gov/pubmed/20576703|GeneMANIA]] * '''We are interested in discussing custom analysis''' - it is how we learn what you need. |
| Line 10: | Line 21: |
| == Veronique Voisin == | * '''What can you expect from the service:''' * We run pathway and network analysis for you and help interpret the data. * We can help you at different stages: * at the experimental design stage * during the analysis: we offer training in data analysis and exploration * after an initial analysis is complete and any validation experiments have been performed, we can book a follow-up meeting to see if you need additional analyses or to help plan subsequent genomics experiments. <<BR>><<BR>> |
| Line 12: | Line 30: |
| veronique.voisin@gmail.com | {{attachment:website2.png}} |
| Line 14: | Line 32: |
| located at TMDT 8th floor each Tuesday | <<BR>> |
| Line 16: | Line 34: |
| ---- ----- |
=== How to use the service === |
| Line 19: | Line 36: |
| == Introduction about the service: == | * '''If you are a member of OICR Cancer Stem Cell program''', you may use the service if * You are planning to generate 'omics' (e.g. gene expression) data * You have a large gene list derived from a large-scale omics project that is ready to be analyzed * You require training in pathway and network analysis |
| Line 21: | Line 41: |
| We are developing a new pathway and network bioinformatics analysis service to help CSC program researchers. High-throughput genomic experiments (e.g. gene expression, protein expression, molecular interactions, large-scale genetic screens and other omics data) often lead to the identification of large gene lists. The interpretation of results and the formulation of consistent biological hypotheses from these gene lists are challenging. Pathway and network analysis can ease the interpretation of this data and formulation of consistent biological hypotheses. ---- ----- |
<<BR>> '''Please schedule an appointment with us:''' * '''Consulting meeting''' If you are planning a genomics experiment and you need some advice concerning the experiment design. Typical time: 30-60 minutes |
| Line 26: | Line 45: |
| == Information about Pathway and Network Analysis == | * '''Analysis planning meeting''' If you have data ready to analyze and they have been already statistically analyzed. Typical time: 60 minutes * '''Training session''' If you want to do your own pathway and network analyses, we can explain how software tools and methods work, such as GSEA, Enrichment Map and GeneMANIA. Typical time: Regular training schedule is currently being planned. Individual or group sessions can be arranged. |
| Line 28: | Line 49: |
| ---- ----- |
* [[CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService/SOP | More details following this link ]] |
| Line 31: | Line 51: |
| == Information about the software we use (software and tutorials) == === GSEA === === Cytoscape === === EnrichmentMap === === WordCloud === === GeneMania === |
{{attachment:flowchart.png|flowchart}} <<BR>> === How to book an appointment === 1. Normal in person meetings are on Tuesdays at TMDT 8th floor. Let us know if this doesn't work for you. [[CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService/Calendar |Check our meeting calendar to see available times]] (30 min to 60 minutes meeting) *Send an e-mail to veronique.voisin@gmail.com with your preferred meeting time and the purpose of the meeting and wait for e-mail confirmation. *For first-time meetings, please send a paper that best describes your work prior to the meeting. *If you booked an initial meeting, please [[CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService/SOP| read our standard operating procedure to know what to expect]] *If you must cancel a meeting, please give 24 hours notice to veronique.voisin@gmail.com. <<BR>> *[[CSCPathwayAnalysisService/Calendar | LINK TO CALENDAR ]] *[[CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService/SOP | LINK TO SERVICE SOP ]] *[[CSCPathwayAnalysisService/Tutorials | LINK TO TUTORIAL PAGE ]] |
| Line 41: | Line 71: |
== Service SOP == ---- ----- == Standard input requirements == ---- ----- == How to use the service == * how to book an appointment * calendar ---- ----- == List of projects == ---- ----- == ? Link to results and reports ? == ---- ----- |
OICR Cancer Stem Cell program - Pathway and Network Analysis Service
The Pathway and Network Analysis Service is freely available to all Cancer Stem Cell program members.
High-throughput genomic experiments (e.g. gene expression, large-scale genetic screens) often lead to the identification of large gene lists. The interpretation of results and the formulation of consistent biological hypotheses from these gene lists can be challenging. Pathway and network analysis (e.g enrichment analysis) approaches can aid interpretation by relating the gene list to knowledge about the biological system, such as pathways.
Our goal is to help researchers interpret results of genomics experiments. Analysis is conducted in close collaboration with researchers on each project to ensure correct input data and effective interpretation of results. Ideally researchers do as much of the analysis and interpretation as they can.
Standard types of pathway analysis offered
Pathway and network analysis: find pathways enriched in a list of genes (e.g. differentially expressed genes)
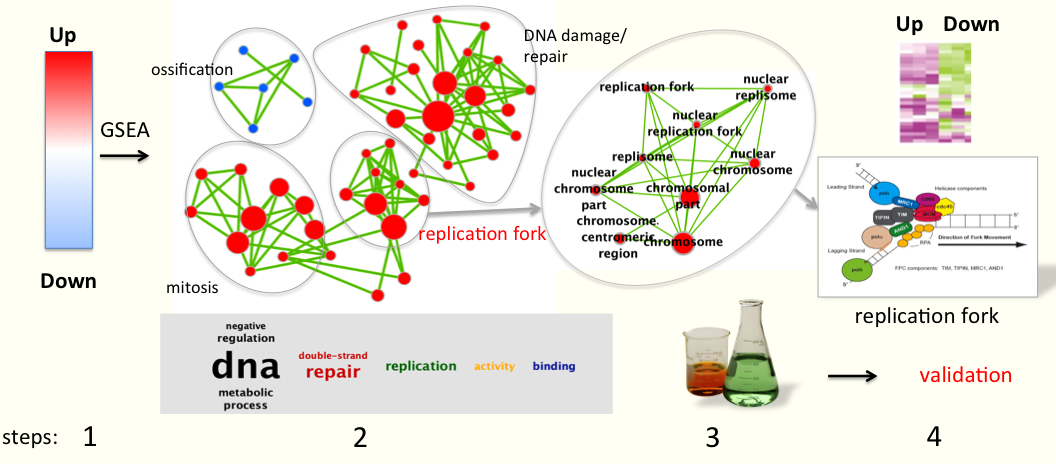
Gene-set enrichment analysis helps characterize large gene lists by finding functionally coherent gene-sets, such as pathways, that are statistically over-represented in a given gene list. We have also developed a method to visualize the results of this analysis, called Enrichment Map. Enrichment Map organizes gene-sets in a network and it enables the user to quickly identify the major enriched functional themes. Input: gene list from genomics experiment (statistically analyzed). Output: enriched pathways visually displayed.
Related publications: Gene-set enrichment analysis (GSEA) Enrichment Map
Predict the function of an unknown gene GeneMANIA finds other genes that are related to a set of input genes, using a very large set of functional association data. Input: a gene or set of genes. Output: connections between input genes and suggestions for additional related genes.
Related publications: GeneMANIA
We are interested in discussing custom analysis - it is how we learn what you need.
What can you expect from the service:
- We run pathway and network analysis for you and help interpret the data.
- We can help you at different stages:
- at the experimental design stage
- during the analysis: we offer training in data analysis and exploration
- after an initial analysis is complete and any validation experiments have been performed, we can book a follow-up meeting to see if you need additional analyses or to help plan subsequent genomics experiments.
How to use the service
If you are a member of OICR Cancer Stem Cell program, you may use the service if
- You are planning to generate 'omics' (e.g. gene expression) data
- You have a large gene list derived from a large-scale omics project that is ready to be analyzed
- You require training in pathway and network analysis
Please schedule an appointment with us:Consulting meeting If you are planning a genomics experiment and you need some advice concerning the experiment design. Typical time: 30-60 minutes
Analysis planning meeting If you have data ready to analyze and they have been already statistically analyzed. Typical time: 60 minutes
Training session If you want to do your own pathway and network analyses, we can explain how software tools and methods work, such as GSEA, Enrichment Map and GeneMANIA. Typical time: Regular training schedule is currently being planned. Individual or group sessions can be arranged.
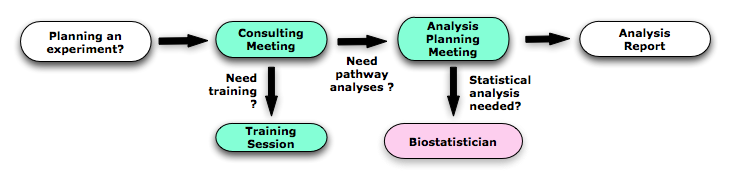
How to book an appointment
Normal in person meetings are on Tuesdays at TMDT 8th floor. Let us know if this doesn't work for you. Check our meeting calendar to see available times (30 min to 60 minutes meeting)
Send an e-mail to veronique.voisin@gmail.com with your preferred meeting time and the purpose of the meeting and wait for e-mail confirmation.
- For first-time meetings, please send a paper that best describes your work prior to the meeting.
If you booked an initial meeting, please read our standard operating procedure to know what to expect
If you must cancel a meeting, please give 24 hours notice to veronique.voisin@gmail.com.