|
Size: 2606
Comment:
|
Size: 4135
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| {{attachment:Software/EnrichmentMap/UserManual/enrichmentmap_logo3.png|Enrichment Map Logo|align="right"}}<<BR>> | |
| Line 6: | Line 6: |
---- ----- == Veronique Voisin == veronique.voisin@gmail.com located at TMDT 8th floor on Tuesday |
|
| Line 26: | Line 17: |
| == Information about Pathway and Network Analysis == | |
| Line 29: | Line 19: |
| All the software used are freely available (open-source). | == How to use the service == === Who can use the service === You can use the service if you are a member of Cancer Stem Cell program, if you are planning to generate omics data, or if you already have large gene lists coming from large-scale 'omics' (e.g. genomics) projects that are ready to be analyzed. Please, book an appointment with us for an initial meeting, a consulting meeting or a training session (see description below). * If you have data ready to analyze and they have been already statistically analyzed, you can book an initial meeting. * If you plan a genomics experiment and you need some advice concerning the experiment design, you can book a consulting meeting. * If you want to do your enrichment analyses on your own, you can book a training session and we can explain you how to use Enrichment Map, Word Cloud and GeneMANIA. {{attachment:flowchart.png}} === How to book an appointment === 1. Look at the calendar below to see my available times the day you want to meet (30 min to 1 h meeting). Be aware of that I'm available for meetings only on Tuesdays! *Send me an e-mail at veronique.voisin@gmail.com to indicate when you want to meet and the purpose of the meeting. Im located at TMDT 8th floor on Tuesday. *I will send you an e-mail back to confirm the appointment. *If we meet for the first time, I encourage you to send me a paper that best describe your work prior to our meeting. *You must cancel a meeting 24 hours in advance. Send an e-mail at veronique.voisin@gmail.com to cancel an appointment. === Data input requirement: please have these data and information ready === [[CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService/SOP | link to SOP ]] ---- ----- ---- ----- == Calendar == <<GoogleCalendar(i6u58gktnv6b3ulah4kmte0me0@group.calendar.google.com, "America/Toronto", 500, 400)>> |
| Line 35: | Line 64: |
| == Online tutorials == === Gene-Set Enrichment Analysis (GSEA) === *http://www.broadinstitute.org/gsea/index.jsp : Gene Set Enrichment Analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g. phenotypes). We use GSEA at the first analysis step. As we will perform this analysis for you, you don"t need to download GSEA. === Cytoscape === * http://www.cytoscape.org/: Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data === EnrichmentMap === * http://baderlab.org/EnrichmentMap === WordCloud === === GeneMania === |
== Information about Pathway and Network Analysis == |
| Line 48: | Line 66: |
| * Suggested readings: * GSEA Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545-50. http://www.ncbi.nlm.nih.gov/pubmed/16199517 * Enrichment Map: Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. Merico D, Isserlin R, Stueker O, Emili A, Bader GD. PLoS One. 2010 Nov 15;5(11):e13984. http://www.ncbi.nlm.nih.gov/pubmed/21085593 |
|
| Line 51: | Line 81: |
| == How to explore an interactive Enrichment Map on your computer == | |
| Line 52: | Line 83: |
| == Service SOP == | [[CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService/Tutorials | link to tutorial page ]] |
| Line 54: | Line 85: |
| ---- ----- |
|
| Line 57: | Line 86: |
| == Standard input requirements == | |
| Line 59: | Line 87: |
| ---- ----- |
|
| Line 62: | Line 88: |
| == How to use the service == * how to book an appointment * calendar <<GoogleCalendar(97aedd6b503d4fd5d4b0f177e0f47f83@group.calendar.google.com, "America/Toronto", 1000, 800)>>---- ----- |
|
| Line 68: | Line 89: |
| == List of projects == | |
| Line 70: | Line 90: |
| ---- ----- == ? Link to results and reports ? == |
Pathway and Network Analysis Service
Cancer Stem Cell program
Introduction about the service:
- The Pathway and Network Analysis Service is freely available to all Cancer Stem Cell program members. High-throughput genomic experiments (e.g. gene expression, protein expression, molecular interactions, large-scale genetic screens and other omics data) lead to the identification of large gene lists. The interpretation of results and the formulation of consistent biological hypotheses from these gene lists are challenging. Computational approaches can aid interpretation by relating the gene lists to knowledge about the biological system. To help researchers interpret their results, we are developing a new consulting and analysis service for pathway and network analysis. Analysis will be conducted in close collaboration with researchers on each project (Cancer Stem Cell research program) to ensure correct input data and effective interpretation of results.
How to use the service
Who can use the service
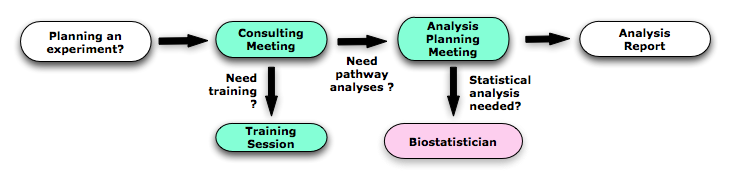
- You can use the service if you are a member of Cancer Stem Cell program, if you are planning to generate omics data, or if you already have large gene lists coming from large-scale 'omics' (e.g. genomics) projects that are ready to be analyzed. Please, book an appointment with us for an initial meeting, a consulting meeting or a training session (see description below).
- If you have data ready to analyze and they have been already statistically analyzed, you can book an initial meeting.
- If you plan a genomics experiment and you need some advice concerning the experiment design, you can book a consulting meeting.
- If you want to do your enrichment analyses on your own, you can book a training session and we can explain you how to use Enrichment Map, Word Cloud and GeneMANIA.
How to book an appointment
- Look at the calendar below to see my available times the day you want to meet (30 min to 1 h meeting). Be aware of that I'm available for meetings only on Tuesdays!
Send me an e-mail at veronique.voisin@gmail.com to indicate when you want to meet and the purpose of the meeting. Im located at TMDT 8th floor on Tuesday.
- I will send you an e-mail back to confirm the appointment.
- If we meet for the first time, I encourage you to send me a paper that best describe your work prior to our meeting.
You must cancel a meeting 24 hours in advance. Send an e-mail at veronique.voisin@gmail.com to cancel an appointment.
Data input requirement: please have these data and information ready
Calendar
Information about Pathway and Network Analysis
- Suggested readings:
- GSEA
- Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545-50.
- Enrichment Map:
- Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. Merico D, Isserlin R, Stueker O, Emili A, Bader GD. PLoS One. 2010 Nov 15;5(11):e13984.
- GSEA
How to explore an interactive Enrichment Map on your computer
