|
Size: 17025
Comment:
|
← Revision 164 as of 2014-09-24 14:23:23 ⇥
Size: 6463
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| #acl CscGroup:read,write,revert | ## page was renamed from CancerStemCellProject/VeroniqueVoisin/PathwayAnalysisService #acl All:read <<BR>> {{attachment:logo.png|OICR_CSC Pathway and Network Analysis Logo Map Logo|align="right"}} = OICR Cancer Stem Cell program - Pathway and Network Analysis Service = |
| Line 3: | Line 7: |
| = Pathway and Network Analysis Service = | '''The Pathway and Network Analysis Service is freely available to all OICR Cancer Stem Cell program members.''' <<BR>> |
| Line 5: | Line 9: |
| == Cancer Stem Cell program == | == Goals of the service == * '''High-throughput genomic experiments''' (e.g. gene expression, large-scale genetic screens) often lead to the identification of large gene lists. The interpretation of results and the formulation of consistent biological hypotheses from these gene lists can be challenging. Pathway and network analysis (e.g enrichment analysis) approaches can aid interpretation by relating the gene list to knowledge about the biological system, such as pathways. |
| Line 7: | Line 12: |
| ---- ----- |
* '''Our goal''' is to help researchers interpret results of genomics experiments. Analysis is conducted in close collaboration with researchers on each project to ensure correct input data and effective interpretation of results. Ideally researchers do as much of the analysis and interpretation as they can. |
| Line 10: | Line 14: |
| == Veronique Voisin == | * We are also focusing on developing training materials and sessions to help researchers who are interested to perform these bioinformatics analyses themselves. Examples of published pathway maps and list of tutorials that could guide researcher can be found following this link: [[CSCPathwayAnalysisService/Tutorials | RESOURCES AND EXAMPLES]]. |
| Line 12: | Line 16: |
| veronique.voisin@gmail.com | == Standard types of pathway analysis offered == |
| Line 14: | Line 18: |
| located at TMDT 8th floor on Tuesday | * '''Pathway and network analysis: find pathways enriched in a list of genes (e.g. differentially expressed genes)''' * Gene-set enrichment analysis helps characterize large gene lists by finding functionally coherent gene-sets, such as pathways, that are statistically over-represented in a given gene list. We have also developed a method to visualize the results of this analysis, called Enrichment Map. Enrichment Map organizes gene-sets in a network and it enables the user to quickly identify the major enriched functional themes. '''Input''': gene list from genomics experiment (statistically analyzed). '''Output''': enriched pathways visually displayed. |
| Line 16: | Line 21: |
| ---- ----- == Introduction about the service: == The Pathway and Network Analysis Service is freely available to all Cancer Stem Cell program members. High-throughput genomic experiments (e.g. gene expression, protein expression, molecular interactions, large-scale genetic screens and other omics data) lead to the identification of large gene lists. The interpretation of results and the formulation of consistent biological hypotheses from these gene lists are challenging. Computational approaches can aid interpretation by relating the gene lists to knowledge about the biological system. To help researchers interpret their results, we are developing a new consulting and analysis service for pathway and network analysis. Analysis will be conducted in close collaboration with researchers on each project (Cancer Stem Cell research program) to ensure correct input data and effective interpretation of results. ---- ----- |
* '''Example of pathway and network analysis:''' [[CSCPathwayAnalysisService/Publication | (MORE EXAMPLES) ]] * A typical example (see figure below the text) comes from gene expression data comparing treated samples versus non-treated samples. The first step is to identify differential gene expression using statistics: genes are ranked using t-test t values with up-regulated genes at the top of the list and down-regulated genes at the bottom. * Next, GSEA is run to find out if gene-sets contain mostly up or down-regulated genes. [Gene-sets are a group of genes that have been annotated to have a similar biological function or belong to the same biological pathway e.g. mitosis and are collected from multiple databases]. * Then, Enrichment map helps visualize all the gene-sets that are significantly enriched in the treated (red circles) or in the non-treated samples (blue circles). [Each gene-set is represented by a circle, also known as a node]. If gene-sets have similar annotations, they cluster together on the map [e.g. all gene-sets related to chromosome condensation and replication fork cluster together] which ease interpretation of the map. In this example, many gene-sets related to mitosis and DNA replication/damage, or involved in the replication fork complex, are enriched in the treated samples (red nodes, genes in these gene-sets are mostly up-regulated). Gene-sets involved in ossification/bone morphogenesis are enriched in the non-treated samples (blue nodes). * As a result, the analysis output summarizes all of the known biological function/pathways that are changing in a particular experiment and more detailed analyses can be performed as a next step to validate or to generate new hypotheses. |
| Line 27: | Line 28: |
| {{attachment:website2.png}} * '''Predict the function of an unknown gene''' GeneMANIA finds other genes that are related to a set of input genes, using a very large set of functional association data. '''Input''': a gene or set of genes. '''Output''': connections between input genes and suggestions for additional related genes. * Related publications: [[http://www.ncbi.nlm.nih.gov/pubmed/20576703|GeneMANIA]] * '''We are interested in discussing custom analysis''' - it is how we learn what you need. == Statistical Analysis == * Pathway and network analysis comes when a gene list has been generated from high throughput OMICs experiments and needs to be functionally interpreted. The data should have then been already statistically analyzed. If your list contains true positives, you are going to be more confident about the output of the pathway analysis. On the other hand, if the gene list contains more noise, we will have to be more cautious about the interpretation of the results and it will also require additional analyses that will delay the overall process of interpretation. Experience is showing us that taking a lot of care in the early steps of the statistical analysis -- by using the statistical method that best fit your data including normalization or removing outliers -- improve the pathway and network analysis results. For these reasons, we have also developed a biostatistics service that can help you if you need to choose a method or process your data in a correct format for subsequent pathway and network analyses: * Please look at http://www.baderlab.org/CSCBiostatService for more information. * You are also encouraged to contact us as soon '''as you plan''' your experiment: genomics technologies can be very sensitive to noise and a well designed experiment is very important for best results. Statistical consultation at the design stage is crucial for improved data quality and results. |
|
| Line 30: | Line 42: |
| === Who can use the service === | * Please follow [[CSCPathwayAnalysisService/HowToUse | THIS LINK ]] to get more details about what to expect from the service and suggested data requirements. |
| Line 32: | Line 44: |
| You can use the service if you are a member of Cancer Stem Cell program, if you are planning to generate omics data, or if you already have large gene lists coming from large-scale 'omics' (e.g. genomics) projects that are ready to be analyzed. Please, book an appointment with us for an initial meeting, a consulting meeting or a training session (see description below). === What you can expect === === How to book an appointment === 1. Look at the calendar below to see my available times the day you want to meet (30 min to 1 h meeting). Be aware of that I'm available for meetings only on Tuesdays! *Send me an e-mail at veronique.voisin@gmail.com to indicate when you want to meet and the purpose of the meeting. *I will send you an e-mail back to confirm the appointment. *If we meet for the first time, I encourage you to send me a paper that best describe your work prior to our meeting. *You must cancel a meeting 24 hours in advance. Send an e-mail at veronique.voisin@gmail.com to cancel an appointment. === Data input requirement: please have these data and information ready === * During the first initial meeting, we are going to discuss : * the biological question(s) you want to answer * the experimental design * the platform you used to generate your data (e.g Affymetrix or Illumina, the chip model,...) * the quality controls and the input data format * Your data should have been statistically analyzed (you should provide us with one file containing this information): * The data should have been normalized. * Some control quality plots should have been done: * Box-plot of intensity (before and after normalization) * Principal Component Analysis (PCA) * Hierarchical clustering of samples (performed on all the data) * Please provide a powerpoint presentation with a figure for each analysis * An appropriate statistical test testing your hypothesis (your biological question) should have been performed * for example : moderated t-test, paired t-test, ANOVA,... * If you need support for your statistical analyses, please contact Shaheena Bashir (Ph.D. in Statistics) at sbashir@uhnres.utoronto.ca. Located at MaRS TMDT 15th floor, she offers free consultation for statistical analyses for Cancer Stem Cell program. She will analyze your data and output the results in the right format for subsequent enrichment analyses. You are encouraged to contact Shaheena as soon as you plan your experiment: these genomics technologies are very sensitive to noise and a well designed experiment is very important for best results. It will ensure better quality data. * You need to provide us with 1 file (.txt) for the enrichment analysis : * Name your file as follow: yourname_date.txt (example: veronique_March21.txt) * Please rename your file with a new date if you resubmit your file * Please follow the format description: * the first column corresponds to Entrez ID. * An Entrez ID is a numerical value that uniquely identifies genes. * For example the Entrez ID for Myc (myelocytomatosis oncogene [ Mus musculus ]) is 17869: http://www.ncbi.nlm.nih.gov/gene/17869. * the second column corresponds to a unique array identifier (ProbesetID for Affymetrix and sampleID for Illumina). * the third column corresponds to gene name (official gene symbol). * the fourth column corresponds to the gene description (full gene name). * the fifth and sixth columns contain the statistical values : * the statistical values are the ones that enable you to tell if a gene is significantly differentially expressed or not, it could be for example the t value and the p-value if you applied a t-test. * the whole table is ordered by the '''absolute value of the fifth column''' ( t value in this example) in a decreasing order. * the additional columns contain the transformed (log2 for example) and normalized (RMA or quantile normalization for example) values for each sample (= each chip if gene expression data). * Example: ||Entrez ID||Probeset ID||Gene Name||Gene Description||t value||p-value||sample1||sample2||sample3|| ||17218||10572906||Mcm5|| minichromosome maintenance deficient 5, cell division cycle||44.0079||0.001||9.13084||9.7166||8.76638|| ||27279||10448307||Tnfrsf12a||tumor necrosis factor receptor superfamily, member 12a||-41.815||0.001||8.58977||9.29698||8.80844|| ||13215||10582809||Tk1||thymidine kinase 1||39.9456||0.001||8.94519||9.56513||8.38612|| ||12937||10384145||H2afv||H2A histone family, member V||-33.6475||0.001||10.574||10.7741||10.5401|| ||207277||10526848||A430033K04Rik||A430033K04Rik||33.3352||0.001||8.25088||8.4121||8.2783|| * Note: Each row of the table should correspond to a different gene. If several rows correspond to the same gene (same Entrez ID), there are 2 possibilities to remove the redundancy: * for a same gene, only the row corresponding to the best t-value is conserved * for a same gene, the average of the different normalized values is calculated before the t-test is applied * the choice has to be made before the statistical data are performed. We can discuss it during the initial meeting. ---- ----- == Service SOP == * ~+ ''' Consulting Meeting''' +~ * ~+Goal+~: to help plan an experiment before it is run. We can recommend case studies that user can learn from. * Genomics technologies are very sensitive: they can detect small amounts of variation. A good experimental design takes into account all possible variables (or factors) and ensure better quality data. You are encouraged to come to talk with us (and/or with the biostatistician Shaheena Bashir)about your planned experiment. Will your design enable to answer your questions? Are there variables you did not think of? Do you have enough replicates? What is your control? * These consulting meetings can also generate a follow up plan, where additional meetings can be scheduled during and after an experiment is run to answer questions and check that the experimental design is good: it is up to you to decide if you need it or not. We are always available to speak with you about your data. * ~+ ''' Initial Meeting ''' +~ (when a dataset is ready to be analyzed) * ~+Goal+~: We will discuss your project, the biological question(s) you want to answer, the experimental design, the enrichment analysis, statistical data data input formats, and create a project name. * Once correct input data are received and the quality controls are good, we will issue an initial pathway analysis plan (see below). * ~+time estimate+~: 30min to 1 hour * ~+ ''' Training session ''' +~ * ~+Goal:+~ You can book a training session if you wish to do your enrichment analysis on your own of if you want to explore the map once we have performed the analysis for you. We will explain you how to install Cytoscape and the different plugins (Enrichment Map, WordCloud and GeneMANIA) on your computer and how to play with your data. * ~+time estimate+~: 30min to 1 hour * ~+ '''Initial Pathway Analysis Plan''' +~ * ~+Goal:+~ A pathway analysis plan is a document that state the different analyses that are going to be performed and a time estimate. We write the pathway analysis plan once correct input data are received. It needs to be sign off by researchers and P.I. We send it to you as a Google document. * A meeting can be scheduled if requested to explain the Pathway Analysis Plan. * ~+ '''Run analysis, interpret the map and produce a report'''+~ * ~+Status :+~ the analysis status will be visible on the website page; We will communicate with you very regularly during the process to ensure effective interpretation of results. * ~+'''Analysis Report'''+~: A report including a global figure of the map and a detailed focus analysis of several pathways as examples will be written at the end of the analysis. * ~+ '''Analysis Report Meeting'''+~ * Goal: discuss the analysis and report. * Examples of questions we can discuss: Do the results meet your expectations? Is there anything unexpected in the results? If you had the resource, which experiments would you conduct based on the results of this analysis? * Time estimate: 30min to 1hour * Two options are available after this meeting: * We need to perform additional bioinformatics analyses : customized analyses * You are satisfied with the map and we let you play with the data and perform some validation experiments before a follow-up meeting * ~+ '''Customized analyses''' +~ * Meeting with Researcher to explain the results of the customized analyses * ~+ '''Follow-up''' +~ * Goal: you may have performed validation experiments or generated new research hypotheses based on your genomics study. You may need to go back and focus on a different aspect of your data. We can help you to re-analyse your data, provide with additional bioinformatics tools or help planned a next genomics experiment. |
== Link to Tutorials == * Please follow [[CSCPathwayAnalysisService/Tutorials | THIS LINK ]] to get some Enrichment Map examples , some tutorial slides, workflows and tips. |
| Line 140: | Line 48: |
| ----- == Calendar == <<GoogleCalendar(i6u58gktnv6b3ulah4kmte0me0@group.calendar.google.com, "America/Toronto", 500, 400)>> ---- ----- == Information about Pathway and Network Analysis == * Suggested readings: * GSEA Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545-50. http://www.ncbi.nlm.nih.gov/pubmed/16199517 * Enrichment Map: Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. Merico D, Isserlin R, Stueker O, Emili A, Bader GD. PLoS One. 2010 Nov 15;5(11):e13984. http://www.ncbi.nlm.nih.gov/pubmed/21085593 ---- ----- == How to explore an interactive Enrichment Map on your computer == === Download the sofware you need (see below for download and tutorial information) === 1. Cytoscape * Enrichment Map plugin * WordCLoud plugin === Explore the Enrichment map using the .cys file that we give you === * Download the .cys file * Put the .cys file in the directory of your choice * In Cytoscape, go to Open, File and browse the directories to locate your file and click Open. *Explore the map * The “Parameters” tab in the “Results Panel” on the right side of the window contains a legend mapping the colours to the phenotypes and displaying the parameters used to create the map (cut-off values and data files). * The “Network” tab in the “Control Panel” on the left lists all available networks in the current session and at the bottom has a overview of the current network which allows to easily navigate in a network even at higher zoom levels by dragging the blue rectangle (the current view) over the network. *Clicking on a node (the circle that represents a gene set) will open the “EM Geneset Expression Viewer” tab in the “Data Panel” showing a heatmap of the expression values of all genes in the selected gene set. *Clicking on an edge (the line between two nodes) will open the “EM Overlap Expression Viewer” tab in the “Data Panel” showing a heatmap of the expression values of all genes both gene sets that are connected by this edge have in common. *If several nodes and edges are selected (e.g. by dragging a selection box around the desired gene sets) the “EM Geneset Expression Viewer” will show the union of all genes in the selected gene sets and the “EM Overlap Expression Viewer” will show only those genes that all selected gene sets have in common. *The “Geneset Summary” tab in the “Results Panel” on the right contains information about which nodes and edges are selected. === Tips === * Click on “View / Show Graphics Details” to see the map details even on low zoom-levels ---- ----- == Online tutorials == All the software used are freely available (open-source) and easy to install on your computer. === Gene-Set Enrichment Analysis (GSEA) === *http://www.broadinstitute.org/gsea/index.jsp : Gene Set Enrichment Analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g. phenotypes). We use GSEA at the first analysis step. As we will perform this analysis for you, you don't need to download GSEA. === Cytoscape === * http://www.cytoscape.org/: Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data Download Cytoscape 2.8.0 from http://www.cytoscape.org/download.html (need to enter, name, institution, e-mail address but no account necessary) Cytoscape tutorial: http://cytoscape.wodaklab.org/wiki/Presentations/Basic === EnrichmentMap === * http://baderlab.org/EnrichmentMap Enrichment Map is a visualization method for gene set enrichment results which helps quickly find general functional themes in genomics data. Enrichment Map works as a plug-in for Cytoscape. To install it, download the zipped file, move it to the Cytoscape plugin directory and unzip it. === WordCloud === * http://baderlab.org/WordCloud WordCloud is a Cytsocape plugin that generates a word tag cloud from a user-defined node selection, summarizing an attribute of choice. It notably eases the interpretation of the Enrichment Map. Download the WordCloud plugin and put the file in the Cytoscape plugin directory, unzip it and put the WordCloud.jar file in the plugin directory. === GeneMANIA === *http://genemania.org GeneMANIA is a free public resource that offers a simple, intuitive web interface that shows the relationships between genes in a list and analyzes and extends the list to include other related genes. You can use GeneMANIA to find new members of a pathway or complex, find additional genes you may have missed in your screen or find new genes with a specific function. You also can use GeneMANIA as a Cytoscape plugin. You can find the GeneMANIA tutorial at: http://genemania.org/pages/help.jsf ---- ----- == List of projects == This section summarizes the current projects, and the analysis status for each project is very regularly updated. You can see progress in the analysis of your project and see the different priorities assigned to each project. ||project ||lab|| data received || data checked; OK for analysis|| GSEA|| First Map|| Analysis report|| additional analysis|| status||priority|| ||EZ01 ||Zacksenhaus|| Feb 22 || Feb 23|| Feb 24|| Feb 25|| -|| -|| writing the report||1|| ||JD02-map1 ||Dick|| - || -|| -|| -|| -|| -|| -||?|| ||JD02-map2 ||Dick|| - || -|| -|| -|| -|| -|| -||?|| ||JD03 ||Dick|| - || -|| -|| -|| -|| -|| -||?|| ||JD04 ||Dick|| - || -|| -|| -|| -|| -|| -||?|| ||JD05 ||Guidos|| - || -|| -|| -|| -|| -|| -||?|| ---- ----- == ? Link to results and reports ? == ---- ----- ------ |
'''Contact''' Dr. Veronique Voisin (Ph.D Biology) veronique.voisin@gmail.com |
OICR Cancer Stem Cell program - Pathway and Network Analysis Service
The Pathway and Network Analysis Service is freely available to all OICR Cancer Stem Cell program members.
Goals of the service
High-throughput genomic experiments (e.g. gene expression, large-scale genetic screens) often lead to the identification of large gene lists. The interpretation of results and the formulation of consistent biological hypotheses from these gene lists can be challenging. Pathway and network analysis (e.g enrichment analysis) approaches can aid interpretation by relating the gene list to knowledge about the biological system, such as pathways.
Our goal is to help researchers interpret results of genomics experiments. Analysis is conducted in close collaboration with researchers on each project to ensure correct input data and effective interpretation of results. Ideally researchers do as much of the analysis and interpretation as they can.
We are also focusing on developing training materials and sessions to help researchers who are interested to perform these bioinformatics analyses themselves. Examples of published pathway maps and list of tutorials that could guide researcher can be found following this link: RESOURCES AND EXAMPLES.
Standard types of pathway analysis offered
Pathway and network analysis: find pathways enriched in a list of genes (e.g. differentially expressed genes)
Gene-set enrichment analysis helps characterize large gene lists by finding functionally coherent gene-sets, such as pathways, that are statistically over-represented in a given gene list. We have also developed a method to visualize the results of this analysis, called Enrichment Map. Enrichment Map organizes gene-sets in a network and it enables the user to quickly identify the major enriched functional themes. Input: gene list from genomics experiment (statistically analyzed). Output: enriched pathways visually displayed.
Example of pathway and network analysis: (MORE EXAMPLES)
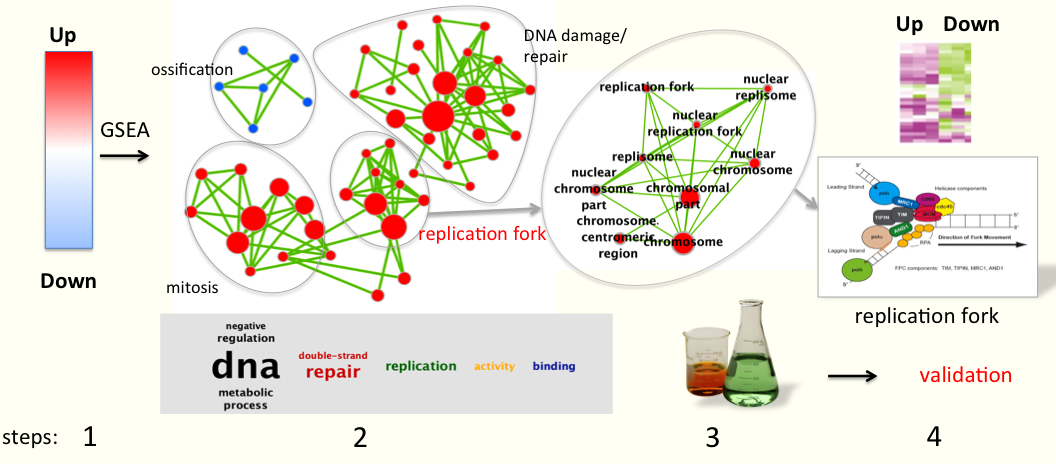
- A typical example (see figure below the text) comes from gene expression data comparing treated samples versus non-treated samples. The first step is to identify differential gene expression using statistics: genes are ranked using t-test t values with up-regulated genes at the top of the list and down-regulated genes at the bottom.
- Next, GSEA is run to find out if gene-sets contain mostly up or down-regulated genes. [Gene-sets are a group of genes that have been annotated to have a similar biological function or belong to the same biological pathway e.g. mitosis and are collected from multiple databases].
- Then, Enrichment map helps visualize all the gene-sets that are significantly enriched in the treated (red circles) or in the non-treated samples (blue circles). [Each gene-set is represented by a circle, also known as a node]. If gene-sets have similar annotations, they cluster together on the map [e.g. all gene-sets related to chromosome condensation and replication fork cluster together] which ease interpretation of the map. In this example, many gene-sets related to mitosis and DNA replication/damage, or involved in the replication fork complex, are enriched in the treated samples (red nodes, genes in these gene-sets are mostly up-regulated). Gene-sets involved in ossification/bone morphogenesis are enriched in the non-treated samples (blue nodes).
- As a result, the analysis output summarizes all of the known biological function/pathways that are changing in a particular experiment and more detailed analyses can be performed as a next step to validate or to generate new hypotheses.
Predict the function of an unknown gene GeneMANIA finds other genes that are related to a set of input genes, using a very large set of functional association data. Input: a gene or set of genes. Output: connections between input genes and suggestions for additional related genes.
Related publications: GeneMANIA
We are interested in discussing custom analysis - it is how we learn what you need.
Statistical Analysis
- Pathway and network analysis comes when a gene list has been generated from high throughput OMICs experiments and needs to be functionally interpreted. The data should have then been already statistically analyzed. If your list contains true positives, you are going to be more confident about the output of the pathway analysis. On the other hand, if the gene list contains more noise, we will have to be more cautious about the interpretation of the results and it will also require additional analyses that will delay the overall process of interpretation. Experience is showing us that taking a lot of care in the early steps of the statistical analysis -- by using the statistical method that best fit your data including normalization or removing outliers -- improve the pathway and network analysis results. For these reasons, we have also developed a biostatistics service that can help you if you need to choose a method or process your data in a correct format for subsequent pathway and network analyses:
Please look at http://www.baderlab.org/CSCBiostatService for more information.
You are also encouraged to contact us as soon as you plan your experiment: genomics technologies can be very sensitive to noise and a well designed experiment is very important for best results. Statistical consultation at the design stage is crucial for improved data quality and results.
How to use the service
Please follow THIS LINK to get more details about what to expect from the service and suggested data requirements.
Link to Tutorials
Please follow THIS LINK to get some Enrichment Map examples , some tutorial slides, workflows and tips.
Contact Dr. Veronique Voisin (Ph.D Biology) veronique.voisin@gmail.com
