Find over represented transcription factor motifs or binding sites in co-expressed genes
- Finding over-representation of transcription factor binding sites in group of genes/pathways found dysregulated in gene expression data
TOOLS THAT USE TRANSCRIPTION FACTOR MOTIFS
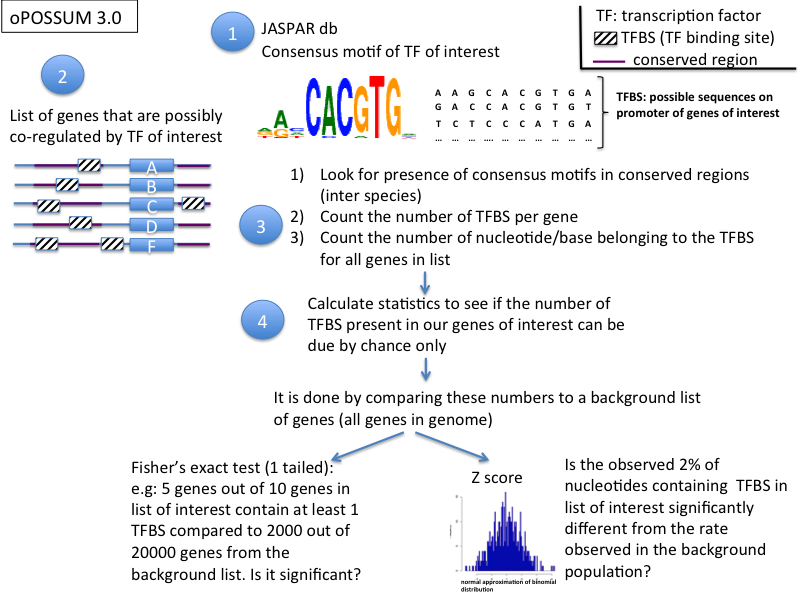
1) oPOSSUM (tested, 3.0) (http://www.cisreg.ca/oPOSSUM/)
- web tool (no account necessary)
- the method has 3 steps:
- phylogenetic footprinting to find regions in the non coding DNA (promoter regions) that are conserved between species
- detection of transcription factor motifs using the JASPAR database (JASPAR PSSMs: position specific scoring matrices)
- 2 statistics methods (Fisher's score and Z score) to evaluate over-represented binding sites compared to background
- tips:
- select potential transcription factor candidates by generating a Z score / Fisher's plot: select the transcription factors that emerged from the cloud
look at the %GC content - Z score to see if you have a %gC content bias: if any, run the GC_compo tool (http://opossum.cisreg.ca/GC_compo/) to select an appropriate background set and use the Sequence-based Single Site Analysis tool
- tip if you use Mozilla Firefox (e.g. 30.0) webbrowser: it can take more than 5 min to get the results back from the oPOSSUM server, in this case you need to change the default response timeout (which is 300s = 5 mins):
- open Firefox
- in the address bar, enter about:config and accept the warning message
- set network.http.response.timeout to 0(no response time out or a time of your choice e.g 3000 for 3000 seconds)
- reopen Firefox and launch oPOSSUM 3.0
2) PSCAN: http://159.149.109.9/pscan/
- need Refseq as gene identifier
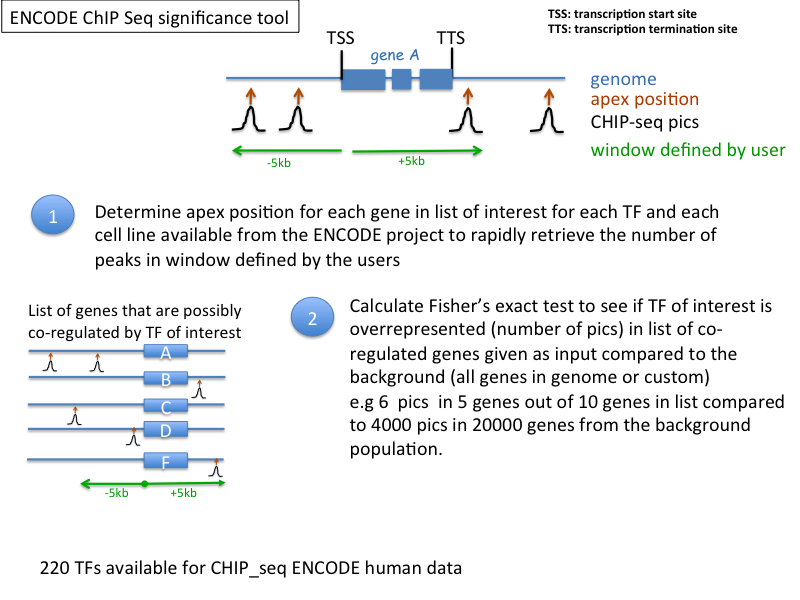
TOOLS THAT USE ENCODE CHIP-seq DATA
ENCODE ChIP-Seq Significance Tool: http://encodeqt.stanford.edu/hyper/
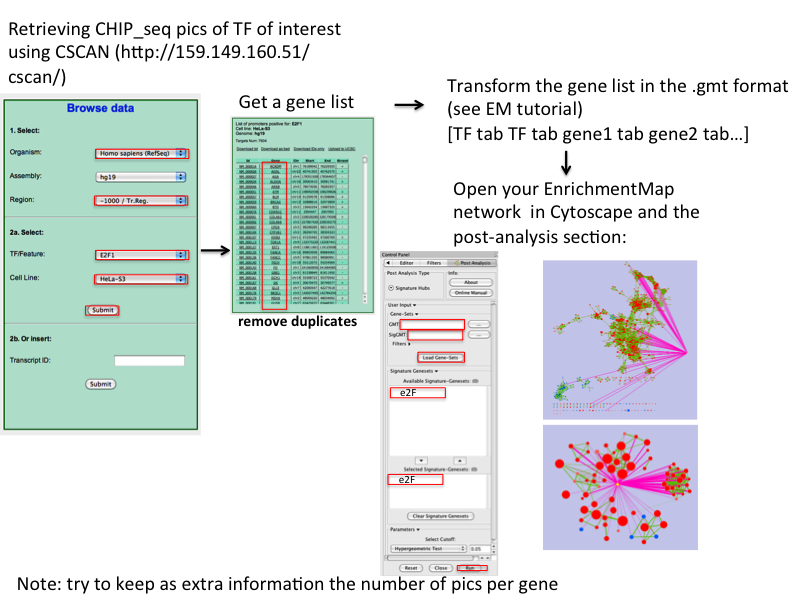
CSAN: (as PSCAn but using chip-seq data): http://159.149.109.9/cscan/
GET THE GENES NEARBY ChIP-SEQ ENCODE DATA and LOOK AT THE MAP OVERLAP using EnrichmentMap POST-ANALYSIS
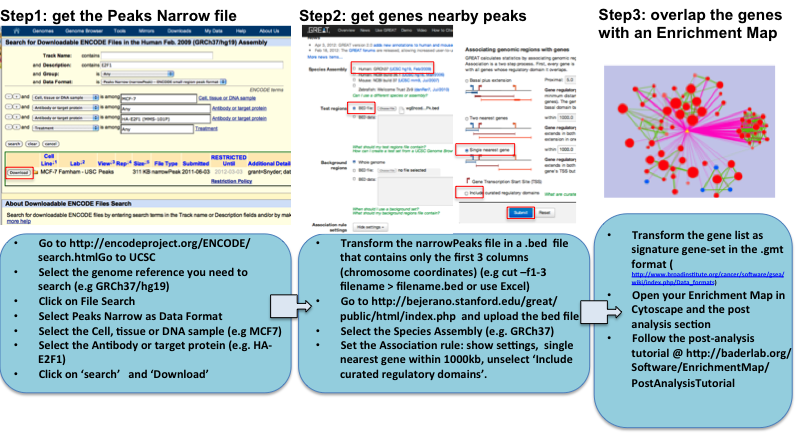
TOOL THAT USES MOTIF DISCOVERY , MOTIF DATABASE and ENCODE CHIP_seq
iRegulon
published in July 2014 : PMID:25058159
tested

- Cytoscape 3 app
- input is a gene list (official gene symbol)
- need to create first a network (see tutorial: import network from table) , then to select nodes (genes) of interest) and run the iRegulon app.
- steps in brief:
- RANKING:
- for each TF motif present in the PWM (position weight matrix) database, rank all genes in the genome by the number of motifs that each gene has on the regulatory region (more exactly: use the Cluster-Buster score for homotypic clusters for each motif). The genes at the top of the ranking are the potential target genes for this motif.
- search for conservation across species
- the Cluster-Buster scores from different motifs corresponding to same TFs are aggregated using a probabilistic method to evaluate the probability of getting same ranking across species by ranking only.
- if multiple motifs refers to same TF, the motif with best rank is kept for the final ranking
- do same ranking with TF CHIP-seq tracks
- RETRIEVED ENRICHED MOTIFS FROM AN INPUT GENE LISTS:
- for the top rank genes (3% by default), an enrichment score is calculated for each rank list (= each motif in the PWM database).
- an Area Under the Curve (AUC) is created by plotting on the x axis the rank list of genes and on the y axis the cumulative recovery of input genes (e.g: 50 genes out of the 100 genes from the input list are found along of the x axis: 50% (0.5) revovery)
- a normalized enrichment score (similar to the GSEA notion) is calculated:
- NES = (AUC value - mean of all AUC rankings) / standard deviation of ALL AUCs
- (NES is a z score indicative of significance if normal distribution)
- NES = (AUC value - mean of all AUC rankings) / standard deviation of ALL AUCs
- LINK MOTIFS TO TRANSCRIPTION FACTORS (motif2TF)
- there are more motifs than transcription factors and sometimes motifs (e/g overrepresented motifs from chip seq data/ de novo motifs discovery) are not yet related to any transcription factors. To increase the number of annotated motifs, they created to network, one based on the sequence homology of the transcription factor and second one based on the homology of the motifs (binding sequence of the TF) and merged the 2 networks. This network is one of the output of the analysis (visualized using Cytoscape)
- RANKING: