Contents
-
A to Z protocol to create an EnrichmentMap from Gene Expression Data and using GSEA (Gene Set Enrichment Analysis)
- System requirement to run this workflow
- Aim and expectations
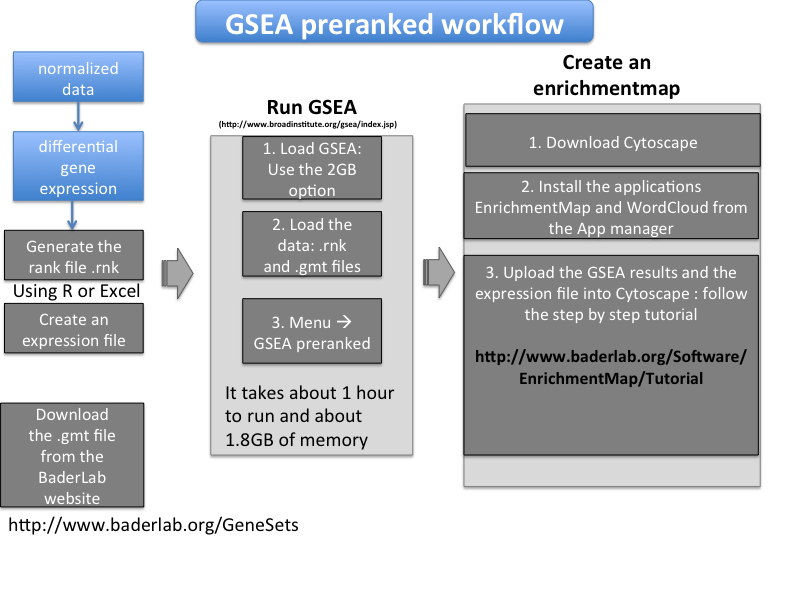
- Sequential steps in brief
- How to create a rank file (.rnk)
- How to get the pathway database file (.gmt)
- How to create your own gene set (e.g to compare your data with a set from another publication) (.gmt)
- How to install GSEA
- How to run GSEA in the preranked mode
- How to create an expression file
- How to create a enrichment map using Cytoscape
- Highlighting the leading edge genes in each gene-set
- What is the next step, how to use the map
-
EXAMPLE 1 WITH AFFYMETRIX MICROARRAY DATA (unpaired analysis)
- description of the data set
- Download the data from GEO
- Install R
- How to preprocess the data (normalization, QC, differential expression)
- How to update the annotations
- How to create a rank file
- How to create an expression file
- How to run GSEA
- How to create a map
- What is the next step, how to use the map
- EXAMPLE 2 CODE FOR AFFYMETRIX MICROARRAYS (paired analysis)
- EXAMPLE 3 CODE FOR ILLUMINA BEADCHIPS (2 grousp comparisons + ANOVA)
A to Z protocol to create an EnrichmentMap from Gene Expression Data and using GSEA (Gene Set Enrichment Analysis)
![]() (DRAFT, UNDER CONSTRUCTION)
(DRAFT, UNDER CONSTRUCTION)
System requirement to run this workflow
- 2GB of free/system memory is needed to run GSEA or navigate through an Enrichment Map. Thus having at least 4GB of system memory is recommended. (you can check it in System Properties on a Windows machine and under About this Mac for mac computers)
2.5 GB is required to build and view an EnrichmentMap with Cytoscape 3.
- 64 bits (you can check it in System Properties on a Windows machine and under About this Mac for mac computers).
Aim and expectations
- The goal of this analysis is to perform a gene set enrichment analysis; it means to look at the genes that are differentially expressed between the 2 (or more) conditions and see if some of these genes belong to same biological function or process. It can be a way of rapidly identifying the major altered biological functions. Doing the analysis at the pathway level is also an efficient way to get over the noise in some dataset: if the differential expression value of a few genes are borderline regarding the significance because of some noise in the data, if the number of these genes belonging to a same biological function is higher than could occur by chance only, taking into account all these expression values for these genes and working at the pathway level could reveal that this pathway is significantly perturbed. The results are represented as a network which has the possibility to add different layers of information in top of each other, making the enrichment results informative.
- The specific goal of this workflow example is to start from gene expression data and to show step by step how to construct and interpret an enrichment map. We would like to know by following this workflow all pathways that could be altered between the 2 (or more) conditions that we are testing. We aim in this analysis to have a global and comprehensive view of what is happening in the cells, a snapshot of the entire cell at the moment the RNA was extracted.
- description of the steps: this section briefly explains the steps that will be followed to run the gene set enrichment workflow as well as the files needed for it.
Sequential steps in brief
- The inital steps are to process the data - your dataset or any data downloadable from the GEO repository - and create files in a correct format for the enrichment analysis. Then the first step of the enrichment analysis will be to run GSEA (Gene Set Enrichment Analysis, which is a tool from the Broad Institute): to do it, we need to create a file called a rank file from the expression data and use it with a pathway database downloadable file from the Baderlab website. Then we will create a network called an enrichment map using the Cytoscape software; for that we will need to create an expression file and use also the GSEA results that we have just run.
How to create a rank file (.rnk)
- the rank file contains only 2 columns: the gene identifiers (official gene symbol in this workflow) as the first column and the differential expression values for each gene as the second column (a fold change (FC, linear or log) or a score [e.g -log10(qvalue) * sign(FC)] or t value ). In this protocol, we will use the t value from a moderated Student's t-test. Headers (column names) should be removed. The format should be tab delimited (meaning that the columns are separated by tabs) and the file extension should be .rnk.
the rank file is a format described in the GSEA documentation: http://www.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats
- the rank file will be used to run the gene set enrichment analysis (GSEA).
 the rank files contains all the genes: do not filter by only genes differentially expressed. We are ranking the genes from the top up regulated to the top down regulated. The genes that do not vary and not of interest are in the middle of the list and GSEA will not look for significance in the middle of the list but the format requirement for GSEA to correctly calculate the statistics is that all genes are listed.
the rank files contains all the genes: do not filter by only genes differentially expressed. We are ranking the genes from the top up regulated to the top down regulated. The genes that do not vary and not of interest are in the middle of the list and GSEA will not look for significance in the middle of the list but the format requirement for GSEA to correctly calculate the statistics is that all genes are listed. example_rank.rnk: it is possible to open this file in a text editor or in excel (e.g. by specifying read 'All Files') to view the format of the file.
How to get the pathway database file (.gmt)
- The pathway database file contains all known and curated biological functions that we are going to test in this pathway enrichment analysis. For each of these functions, the names of genes known to be implicated in this function are listed beside this function. The gene set enrichment analysis will look if the top differentially expressed genes are included in some of these pathways. It will also assess if this enrichment can happen by chance only or not.
In this protocol we are going to use a file that include pathways from different sources (e.g Gene Ontology, Reactome, Kegg, Msigdb c2, ...) and that is updated monthly. We observed that having the most comprehensive set of pathways gave more sensitive results (more info at http://www.baderlab.org/GeneSets). Although databases that are included in this file can be overlapping, they are not 100% identical and the clusters created by the enrichment map from these different sources add confidence about the perturbation of a given biological function.
The link to the database file compiled by the BaderLab and updated monthly can be found at: http://download.baderlab.org/EM_Genesets/ (look for the current release at the bottom of the list) and a description of how the file is being created at: http://baderlab.org/GeneSets
example of file for mouse data (please look for the current release):Mouse_GOBP_AllPathways_no_GO_iea_August_20_2014_symbol.gmt
example of file for human data (please look for the current release):Human_GOBP_AllPathways_no_GO_iea_August_20_2014_symbol.gmt
How to create your own gene set (e.g to compare your data with a set from another publication) (.gmt)
- get the specific gene signature (usually from the supplemental data of publication) that you wish to compare with.
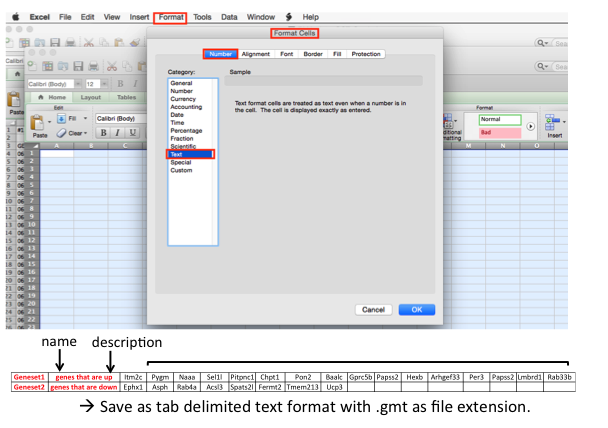
open a new Excel worksheet and format the cells of the worksheet as text (in the menu bar --> Format --> cells)
- copy the gene list and paste / special / transpose in the first row leaving the first 2 columns empty.
- each line can be a different gene signatures.
- save the file as a tab delimited format and use .gmt as file extension
- the .gmt file can be uploaded into GSEA and can replace the pathway database file when GSEA is run.
- recommended length of a gene-set is 250 genes (or a range between 150 and 350).
How to install GSEA
GSEA can be downloaded from http://www.broadinstitute.org/gsea/index.jsp
- you need to enter a valid e-mail address before going to the download section
- for this workflow example, we are going the java web start option to run GSEA: choose the 'launch with 2GB' in the 'javaGSEA Desktop Application' box:

- you may be able to save the icon on your computer (a file called gsea.jnlp). Each time you want to run GSEA, you just need to double-click on the icon. Each time you double click on the icon, GSEA will double check whether a new version if available and install the software in a temporary location in your computer (thus you need a working internet connection to lauch GSEA this way)
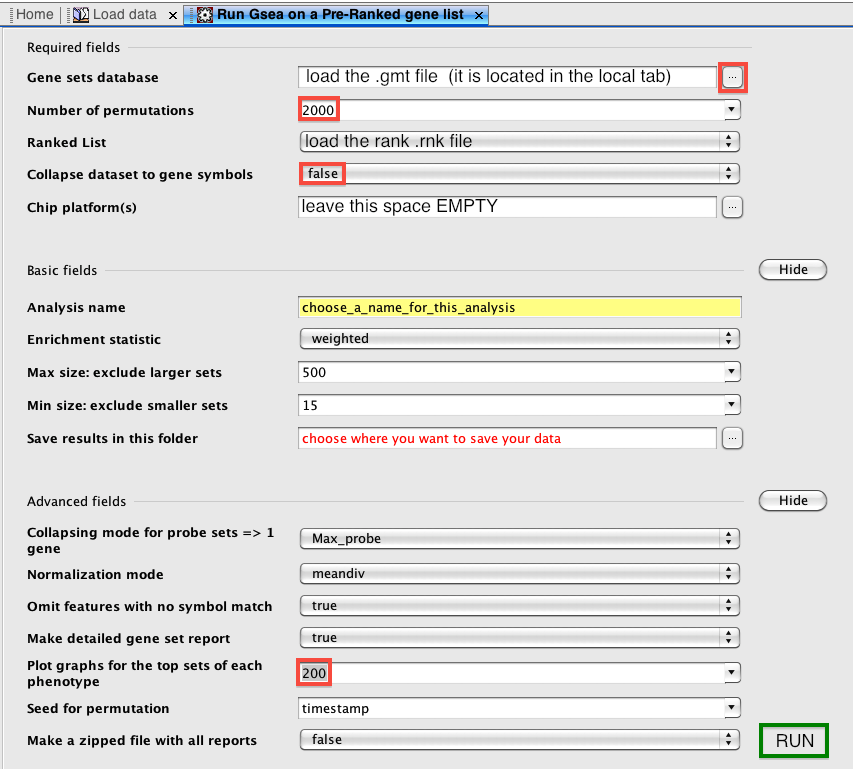
How to run GSEA in the preranked mode
The first step when the application is open is to load the data by browsing or dragging and dropping the files: the .rnk and the .gmt files. Then, it is to open the GSEAPreranked window: menu bar --> Tools --> GseaPreranked. From this window, we typically upload the .rnk and .gmt file (.gmt file is located in the 'Genematrix (local gmx/gmt)' tab). The number of permutations is set to 2000 (1000 or 2000). I find it handy to save the best 200 plots instead of the best 20 plots. Other parameters can be left as default (alternatively, the weight p2 can be used to put a larger weight than default on the top ranked genes, if we wish to increase privilege to most highly differentially expressed genes genes over less significant ones).
How to create an expression file
- Although only 2 files were needed to run GSEA, a additional file, called the expression file,needs to be prepared to create an enrichmentmap. For this workflow example, the expression file will contain as first column the official gene symbol, as second column the full names of the genes (called sometimes definition), followed by columns containing the normalized data for each of the samples included in the study. It is a tab delimited file and the extension can be a .txt file and column names remain.
How to create a enrichment map using Cytoscape
- Once GSEA has been run successfully, a quick check at the quality of the GSEA plots is recommended as well as a look at the number of significantly enriched pathways.
The next step is to create an enrichment map using the Cytoscape software and the EnrichmentMap application.
- We will use Cytoscape 3.1.1 for this workflow example.
Download Cytoscape 3.1.1 from http://www.cytoscape.org/ (you need to enter a valid e-mail address)
Once Cytoscape is open, install EnrichmentMap from the App manager : menu bar --> App Manager --> Install Apps --> type "EnrichmentMap" and click on "Install".
A step by step guided tutorial on how to create a map from GSEA result can be found at: http://www.baderlab.org/Software/EnrichmentMap/Tutorial (step2)
- You will need to upload the GSEA results and the .gmt file (prefer the automatic upload option using the .rpt file: to use the option, do not move the GSEA folder once the analysis has been done). You will also need to upload the expression file in the expression box. A typical threshold for gene expression data would a FDR (q value) set to 0.1.
- We will use Cytoscape 3.1.1 for this workflow example.
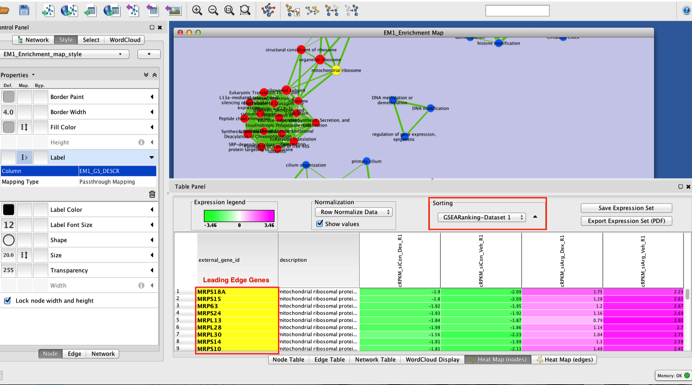
Highlighting the leading edge genes in each gene-set
- Once the map has been built, in Table Panel, make sure that "Sorting" is set to "GSEARanking-Dataset1". It will order the genes from highest differential expression to lower differential expression and you can now see genes labelled in yellow. These genes are the leading edge genes contributing to the enrichment score for each gene-set.
What is the next step, how to use the map
- Once the map is created, it is helpful to play with the q value cut-off to look at the most significant gene-sets, to browse the map and look at the top genes in the significant pathways (in result panel, the sorting option should be set to GSEA ranking-dataset 1). Then the question that should be tried to be answered is: with your knowledge of the model and your specific research question in mind, which pathways were expected and which ones were not? What does it tell us about the biology of the cells that we are studying? Are some pathways more interesting than other ones?
- How to create a figure
- How to interpret the results
What's next? (add different layers of information, use the post-analysis option of EnrichmentMap, create a gene-gene network using GeneMANIA)
- (How to preprocess the data using R)
- (How to preprocess the data using Excel)
EXAMPLE 1 WITH AFFYMETRIX MICROARRAY DATA (unpaired analysis)
description of the data set
Estradiol-treated MCF7 cells, 12 and 24hrs, Gene Expression Omnibus: GSE11352
overall design: "We used oligonucleotide expression microarrays (Affymetrix GeneChip U133 Plus 2.0) to identify estradiol (E2)-responsive genes in the estrogen-receptor positive breast cancer cell line, MCF7. MCF7 cells were grown to 30-50% confluency and exposed to 10 nM E2 (or vehicle only) at 12, 24, and 48 hours. Each timepoint was performed in triplicate (ie, biological replicates). Total RNA was isolated from cells using the Qiagen RNeasy kit, and 5 micrograms of total RNA was amplified, labeled and hybridized to the array according to the manufacturer’s protocols."
Download the data from GEO
Install R
1) install R (http://www.r-project.org/)
2) install RStudio (http://www.rstudio.com/)
3) Go through on online R tutorial (e.g. this one: http://www.cyclismo.org/tutorial/R/)
How to preprocess the data (normalization, QC, differential expression)
How to update the annotations
How to create a rank file
How to create an expression file
How to run GSEA
How to create a map
What is the next step, how to use the map
EXAMPLE 2 CODE FOR AFFYMETRIX MICROARRAYS (paired analysis)