|
Size: 1377
Comment:
|
Size: 1754
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 11: | Line 11: |
| * [[ChangjiangXu/Intranet/Weekly | Weekly Updates]] | |
| Line 18: | Line 17: |
| * [[CancerStemCellProject/ChangjiangXu/HALT02 | HALT02_part2/JAK2]] | * HALT02/JAK2 * [[CancerStemCellProject/ChangjiangXu/HALT02_SRC | Differential expression analysis of SRC high vs low (negative) sub-populations]] * [[CancerStemCellProject/ChangjiangXu/HALT02_JAK2 | Comparison of the pathways for XG-R/PF-R, XG-NR/PF-NR, and XG-R/PF-NR]] |
| Line 22: | Line 23: |
* [[CancerStemCellProject/VeroniqueVoisin/BiostatisticProjects | Potential Biostatistics Projects from Bioinformatics Service]] |
|
| Line 26: | Line 25: |
| * [[CSCBiostatService/AffymetrixDataAnalysis | Affymetrix microarray data analysis]] * [[CSCBiostatService/IlluminaChipDataAnalysis | Illumina’s BeadChip microarray data analysis]] * [[CSCBiostatService/RNAseqDataAnalysis | RNA-seq data analysis]] * [[CSCBiostatService/flowCytometryDataAnalysis | Flow cytometry data analysis]] |
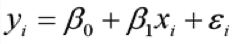
OICR CSC Statistical Analysis Service
OICR Cancer Stem Cell program
Changjiang Xu, Gary Bader
- OICR CSC Projects (PI's name and/or project title listed below)
Laurie Ailles, Crosstalk between CAFs and ovarian cancer cells (LA02_CAF_ovarian_Laurie)
Peter Dirks, Role of dopamine D4 receptor in glioblastoma stem cells
- HALT02/JAK2
Bret Pearson, Spatial Organization of the planarian transcriptome
Bret Pearson, Mechanisms driving asymmetric cell fate outcomes in planarian adult stem cell lineages
