
Enrichment Map Plugin Download Page
Contents
Brief Description
Enrichment Map is a Cytoscape plugin for functional enrichment visualization. Enrichment results have to be generated outside Enrichment Map, using any of the available methods. Gene-sets, such as pathways and Gene Ontology terms, are organized into a network (i.e. the "enrichment map"). In this way, mutually overlapping gene-sets cluster together, making interpretation easier. Enrichment Map also enables the comparison of two different enrichment results in the same map.
Follow this link for more details and an analysis example.
Legends
For easier figure creation download our template legends that contains all the visual properties used in a standard enrichment map. Download and customize legend to fit your network
Plugin Download
Latest Release
Cytoscape 3.4 and higher require Java 8
Download the latest release from cyotscape app store.
For pre-release features download from github.
EnrichmentMap 3.0 is a major release, with the following new features:
EnrichmentMap networks can now be created from any number of enrichment data sets (EnrichmentMap 2.0 supported maximum 2 data sets).
- New chart visualizations on nodes for visualizing NES scores, p-values or q-values. Three new charts are available: radial heat-map, linear heat-map and heat-strips.
- New network creation dialog has the ability to scan a folder for files belonging to each dataset. In most cases this removes the need to manually enter the required files.
- New control panel allows the contents and style of the network and charts to be updated dynamically.
- New legend dialog.
New streamlined HeatMap panel has the ability to summarize expression data.
Several new commands that allow EnrichmentMap to be automated from external scripts and CyREST.
Cytoscape 3
Cytoscape 3.2 and higher require Java 7.
If you are on Mac OSx Snow leopard (10.6.8) and want to use Cytoscape 3.2 check out these instructions for installing Java 7 and Cytoscape 3.2 on your system.
Updated at 11/20/2014 - Release 2.0.1 : Compatible with cytoscape 3.1
- Ported existing EM to Cytoscape 3.
Fixed post analysis (issue #48)
Enrichment Map Plugin (v2.0.1 Build 480)
Download the zipped file, unzip the file, and install the .jar file from file in App manager.
Source code can be found on github: https://github.com/BaderLab/EnrichmentMapApp (EM_Cyto_port branch)
Development Versions
Updated at 05/01/2016 - Release 2.1.0 (Release Candidate 3) : Compatible with cytoscape 3.2
New auto annotation feature. - Check out tutorial here
New Post Analysis options and features including Mann-Whitney post analysis edge calculation.
- fixed issue with renaming clusters.
- fixed null pointer with heatmap creation
- Added one sided mann-whitney test for post analysis
fixed issue #56, #45, #46, #52, and more...
Enrichment Map Plugin (v2.1.0 Release Candidate 1 Build 568)
Download the zipped file, unzip the file, and install the .jar file from file in App manager (menu bar of the Cytoscape software).
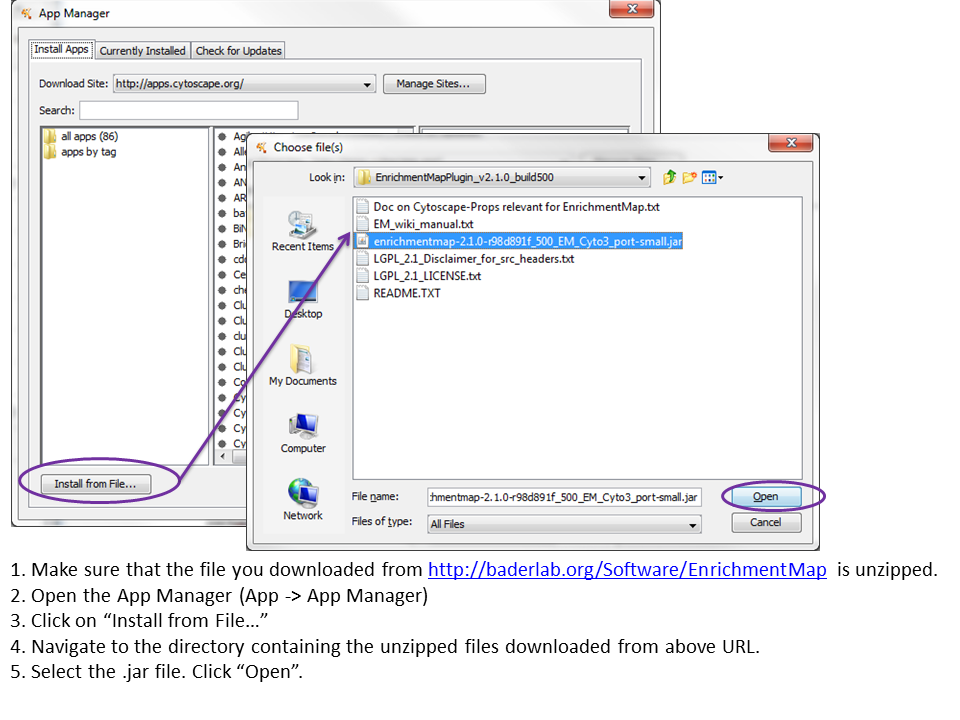
Source code can be found on github: https://github.com/BaderLab/EnrichmentMapApp (AutoAnnotate branch)
Cytoscape 2.8
Updated at 07/06/2011 - Release 1.2 : Compatible with cytoscape 2.6,2.7,2.8
Enrichment Map Plugin (v1.2 Release)
Download the zipped file, move it to the Cytoscape plugin directory and unzip it.
Distributions with source code:
Enrichment Map Plugin with source code, tar and gzipped (v1.2 Release)
Updated at 02/7/2011 - Release 1.1: Compatible with cytoscape 2.6,2.7,2.8
Enrichment Map Plugin (v1.1 Release)
Download the zipped file, move it to the Cytoscape plugin directory and unzip it.
Distributions with source code:
Enrichment Map Plugin with source code, tar and gzipped (v1.1 Release)
Development Versions
Updated at 04/9/2014 - Release 1.3 - Release Candidate 6 : Compatible with cytoscape 2.6,2.7,2.8
- Added bug fix with missing leading edge on reloaded sessions files for down regulated gene sets.
- Added command tool support
- Added EDB support
- Added values to heat map
- Added ability to load GSEA class files directly
- fixed misc bugs
Enrichment Map Plugin (v1.3 RC6)
Download the zipped file, move it to the Cytoscape plugin directory and unzip it.
Source code can be found on github: https://github.com/BaderLab/EnrichmentMapApp
Tutorials and Examples
data used for the tutorials: Estradiol-treated MCF7 cells, 12 and 24hrs (Gene Expression Omnibus: GSE11352)
GSEA format:
Guided GSEA tutorial to load the filed into Enrichment Map
Guided GSEA/EM tutorial for multiple analysis for Cytoscape 3 to create EM directly from GSEA interface UPDATED for Cyotscape 3.
Guided GSEA/EM tutorial for multiple analyses for Cytoscape 2 to create an EM directly from GSEA interface for multiple analysis
Generic format:
- note: the results should be the same as GSEA
DAVID format:
files: EM_David_testData.zip
Guided DAVID Tutorial to load the files into Enrichment Map
BiNGO format:
files: EM_Bingo_testData.zip
Guided BiNGO Tutorial to load the files into Enrichment Map
g:Profiler format:
files: EM_gProfiler_testData.zip
Guided g:Profiler Tutorial to load the files into Enrichment Map
GREAT format
Guided GREAT Tutorial to load files into Enrichment Map
Query-set analysis (aka Post-analysis)
Guided Post Analysis Tutorial to load the files into Enrichment Map
Automatically annotating clusters
User Guide
Gene-sets for Enrichment Analysis
directly downloading our monthly updated gene-set collections from Baderlab genesets collections. Description of sources and methods used to create collection can be found here
- we generate gene-set files for enrichment analysis (e.g. GSEA) by querying public resources such as Gene Ontology and KEGG, and using Entrez-Gene ID for genes
how to convert DAVID gene-sets to GMT: R script
Publications
Cite EM
Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation
Merico D, Isserlin R, Stueker O, Emili A, Bader GD
PLoS One. 2010 Nov 15;5(11):e13984
PubMed Abstract -PDF
Examples of Use
Functional impact of global rare copy number variation in autism spectrum disorders.
Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, et al.
Nature. 2010 Jun 9 (Epub ahead of print)
PubMed Abstract - PDF
Nature Blogs - In the news (The Globe and Mail)
Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps
Isserlin R, Merico D, Alikhani-Koupaei R, Gramolini A, Bader GD, Emili A.
Proteomics 2010, March 10(6):1316-27
Pubmed Abstract - PDF
Papers Citing Enrichment Map
Pathway analysis of expression data: deciphering functional building blocks of complex diseases.
Emmert-Streib F, Glazko GV.
PLoS Comput Biol. 2011 May;7(5):e1002053.
PubMedInflammasome is a central player in the induction of obesity and insulin resistance.
Stienstra R, van Diepen JA, Tack CJ, Zaki MH, van de Veerdonk FL, Perera D, Neale GA, Hooiveld GJ, Hijmans A, Vroegrijk I, van den Berg S, Romijn J, Rensen PC, Joosten LA, Netea MG, Kanneganti TD.
Proc Natl Acad Sci U S A. 2011 Aug 29.
PubMedDelineation of Two Clinically and Molecularly Distinct Subgroups of Posterior Fossa Ependymoma
Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, Benner A, Hielscher T, Milde T, Remke M, Jones DTW, Northcott PA, Garzia L, Bertrand KC, Wittmann A, Yao Y, Roberts SS, Massimi L, Van Meter T, Weiss WA, Gupta N, Grajkowska W, Lach B, Cho YJ, von Deimling A, Kulozik AE, Witt O, Bader GD, Hawkins CE, Tabori U, Guha A, Rutka JT, Lichter P, Korshunov A, Taylor MD, Pfister SM
Cancer Cell, Volume 20, Issue 2, 143-157, 16 August 2011
PubMed Abstract - PDF
Report a Bug or a Problem
please make sure you don't have formatting issues (have a look at the User Guide (and FAQ))
- if you are still not sure how to handle formats,
- or you don't know what's the best suitable analysis for you,
- please send an email to: daniele[AT]merico[DOT]gmail.com
- please check what's your
- plugin version and build (from the Cytoscape menu / Plugins / Enrichment Map / About)
- Cytoscape version (from the Cytoscape menu / Cytoscape)
- Operating System (e.g. Windows Vista)
and send your bug report to ruth[DOT]isserlin[AT]utoronto.ca
OR
report the bug to the Enrichment map issue tracker:- click on "Issues"
- click on "New Issue"
- write a short description of the issue
- attached session file (.cys) file or example input files if applicable
- Make sure to enter plugin version and build, cytoscape version and operating system.
- click on "Submit new issue"
For Feature requests please send to enrichmentmap-dev@googlegroups.com